Class 12th Biology - Biotechnology: Principles and Processes Case Study Questions and Answers 2022 - 2023
By QB365 on 08 Sep, 2022
QB365 provides a detailed and simple solution for every Possible Case Study Questions in Class 12 Biology Subject - Biotechnology: Principles and Processes, CBSE. It will help Students to get more practice questions, Students can Practice these question papers in addition to score best marks.
QB365 - Question Bank Software
Biotechnology: Principles and Processes Case Study Questions With Answer Key
12th Standard CBSE
-
Reg.No. :
Biology
-
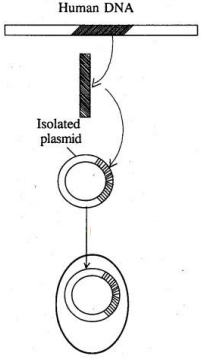
(a) Name the particular technique in Biotechnology, whose steps are shown in the figure.
(b) Name the steps 1 to 4 marked in the figure.
(c) Give an example where a human gene product is. obtained from transgenic bacteria.(a) -
(a) Name and write the significance of enzyme X.
(b) Name and write the role of enzyme Y.
(c) Name the processes A and B.(a) -
Restriction enzymes typically recognize a symmetrical sequence of DNA.
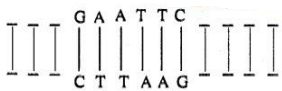
Notice that the top strand is the same as the bottom strand, but reads backward. When the enzyme cuts the strand between G and A, it leaves overhanging chains:

(a) What is this symmetrical sequence of DNA known as?
(b) What is the significance of these overhanging chains?
(c) Name the restriction enzyme that cuts the strand between G and A.(a) -
The following diagram illustrates the linking of DNA fragements. Answer the questions that follow the illustration.
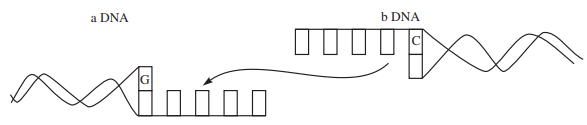
(a) Name 'a' and 'b'.
(b) Complete the palindrome, which is recognised by EcoRI.
(c) Name the enzyme that can link the two DNA fragments.(a) -
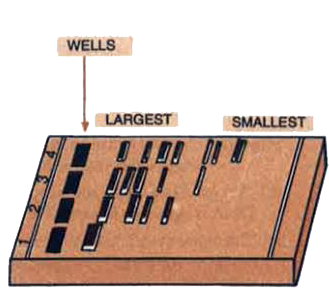
(a) What does the diagram given above, depict?
(b) What is meant by 'largest' and 'smallest' in the diagram'?
(c) Name the compound used to visualise them.
(d) Define elution.(a) -
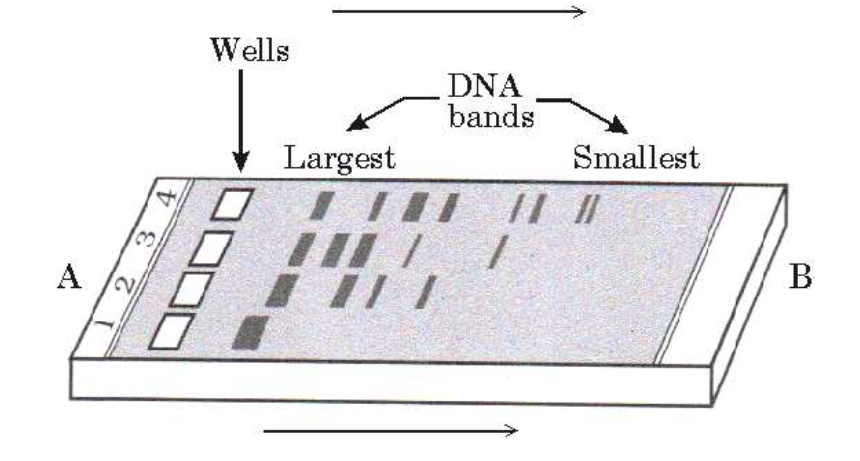
A picture of gel electrophoresis showing the separation of DNA fragments is shown above. Answer the following questions.
(a) Identify the positive and negative terminals.
(b) What is the matrix used and mention its source?
(c) Identify the undigested DNA.(a) -

The E.coli cloning vector pBR322 is shown above.
(a) Identify and name two selectable markers.
(b) Identify and name the restriction enzymes that can be used to ligate an alien DNA fragment at the tetracycline gene.
(c) Identify EcoRI.
(d) Name the gene that controls the copy number of the vector.(a) -
A schematic representation of the steps in Polymerase Chain Reaction (PCR) is shown below. Answer the questions that follow:
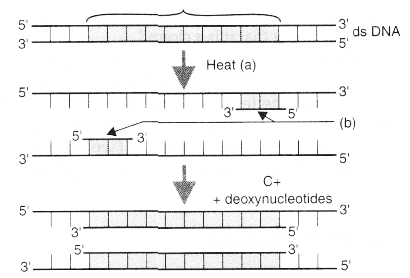
(a) Name the steps A and D in the PCR.
(b) Identify B. What are they chemically?
(c) What is C? Name its source organism.(a) -
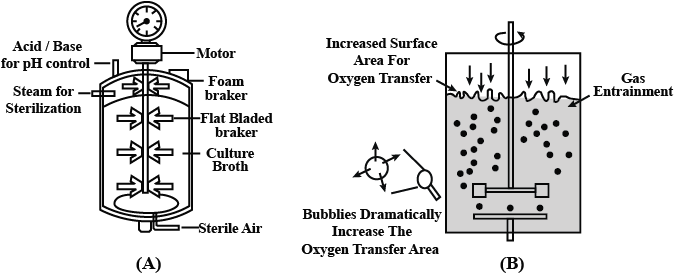
(a) Name the type of bioreactor shown above.
(b) Why does it have a curved base?
(c) What is the significance of the stirrer?(a) -
When a gene product is required in large quantities, the transformants with N2 the plasmid inside the cell are cultured in a large scale in an industrial fermenter, which then synthesise the desired protein. This product is extracted from the fermenter for commercial use.
(a) Why is the used medium drained out from one side while the fresh medium is added from the other. Explain.
(b) List any four optimum conditions for achieving the desired product in a bioreactor.(a) -
Read the following and answer any four questions from (i) to (v) given below:
Gene manipulation is a fast emerging science. It started with development of recombinant DNA molecule. It is named variously as DNA manipulation biotechnology, recombinant DNA technology and genetic engineering. This technology, that mostly involves cutting and pasting of desired DNA fragments, is based on two important discoveries in bacteria, i.e., presence of plasmid in bacteria and restriction endonucleases. Paul Berg was able to introduce a gene of SV-40 into a bacterium. The science of recombinant DNA technology took birth when Cohen and Boyer (1973) were able to introduce a piece of gene containing foreign DNA into plasmid of E.coli.
(i) Biotechnology is also known as(a) DNA manipulation biotechnology (b) recombinant DNA technology (c) genetic engineering (d) all of these. (ii) A bacterial plasmid is a/an
(a) extra chromosomal material that do not replicate (b) extra chromosomal material that undergo replication with or without chromosomal DNA (c) tubular structures that help in conjugation (d) bristle like solid structure that help in adhesion. (iii) Father of genetic engineering is
(a) Paul Berg (b) Arber (c) Nathan (d) Smith. (iv) Which of the following is used by Paul Berg to introduce a gene of SV-40 in a bacterium?
(a) E. coli (b) cos-plasmids (c) Lambda phage (d) None of these (v) Read the given statements and select the correct option.
Assertion : Biotechnology started with the development of recombinant DNA molecule.
Reason : Biotechnology mostly involves cutting and pasting of desired DNA fragments.(a) Both assertion and reason are true and reason is the correct explanation of assertion. (b) Both assertion and reason are true but reason is not the correct explanation of assertion. (c) Assertion is true but reason is false. (d) Both assertion and reason are false. (a) -
Read the following and answer any four questions from (i) to (v) given below:
Restriction endonuclease was isolated for the first time by W Arber in 1962 in bacteria. Restriction endonucleases cut the DNA duplex at specific points therefore they are also called as molecular scissors or biological scissors. Three types of restriction endonucleases are Type I, Type II and Type III but only Type II restriction endonucleases are used in recombinant DNA technology. Restriction endonuclease EcoR I recognises the base sequence GAATTC in DNA duplex and cut strands between G and A.
(i) Only type II restriction enzymes are used in gene manipulation because(a) ATP is not required for cleaving (b) it consists of three different subunits (c) it makes cleavage or cut in both the strands of DNA molecule (d) both (a) and (c). (ii) Which of the following ions are used by restriction endonucleases for restriction?
(a) Mg2+ ions (b) Mn2+ ions (c) Na+ ions (d) K+ ions (iii) Restriction endonuclease was isolated for the first time in a
(a) plant cell (b) animal cell (c) prokaryotic cell (d) germinal cell (iv) Restriction endonucleases are also called as molecular or biological scissors because
(a) they cleave base pairs of DNA only at their terminal ends (b) they cleave one or both the strands of DNA (c) they act only on single stranded DNA (d) none of these (v) Select the option that correctly states the working action of restriction endonuclease EcoR 1 on DNA sequence GAATTC
↓ ↓ ↓ ↓ (a) 5' - GAATTC - 3' (b) 5' - GAATTC - 3' (c) 5' - GAATTC - 3' (d) 5' - GAATTC - 3' 3' _ CTTAAG - 5' 3' - CTTAAG - 5' 3' - CTTAAG - 5' 3' - CTTAAG - 5' ↑ ↑ ↑ ↑ (a) -
Read the following and answer any four questions from (i) to (v) given below:
Tools used in the formation of recombinant DNA are of three types. These are enzymes, cloning vectors and competent host. Lysing enzymes are used to extract DNA for experimental purpose from the cells. Cleaving enzymes break the DNA molecules. They are of three types : exonucleases, endonucleases and restriction endonucleases. A competent host is required for transformation with recombinant DNA and cloning vectors help to propagate DNA.
(i) Which of the following is an example of natural lysing activity in a human body?(a) Lysozyme present in tears dissolve the bacterial cell wall. (b) Conversion of starch to maltose in the buccal cavity (c) Absorption of digested food into the intestinal cells. (d) Conversion of protein molecules into amino acids in the stomach. (ii) The enzyme which has polymerizing activity in 5'→3' direction but exonuclease activity in 3'→ 5' direction only is
(a) RNA polymerase III (b) DNA polymerase II (c) DNA polymerase I (d) All of these (iii) Cloning vectors are the DNA molecules that
(a) carry foreign DNA segment but do not replicate inside the host cell (b) carry foreign DNA segment and replicate inside the host cell (c) transfer nuclear DNA form nucleus to the cytoplasm of the same cells (d) help in sealing gaps in DNA segments. (iv) Transfer of DNA into a eucaryotic cell is called
(a) transformation (b) transduction (c) transfection (d) electroporation. (v) Assertion: Type I restriction enzymes are not used in rDNA technology.
Reason: Type I restriction endonucleases consist of two different subunits and require ATP for restriction activity.(a) Both assertion and reason are true and reason is the correct explanation of assertion. (b) Both assertion and reason are true but reason is not the correct explanation of assertion. (c) Assertion is true but reason is false. (d) Both assertion and reason are false. (a) -
Read the following and answer any four questions from (i) to (v) given below:
The foundations of recombinant DNA (rDNA) were laid by the discovery of restriction enzymes. These enzymes are present in many bacterias where they function as a part of their defense mechanism called the Restriction Modification system (RM system). Molecular basis of this system was explained first by Werner Arber in 1962. The Restriction t-'l0dification system consists of two components:
1. A restriction enzyme (called restriction endonuclease) identifies the introduced foreign DNA and cuts it into pieces.
2.The second component is a modification enzyme (methylase) that adds a methyl group to DNA at specific site to protect it from the restriction enzyme cleavage.
(i) Restriction endonucleases are enzymes present in (i) where they function as a part of (ii) mechanism.(a) (i) bacteria (ii) digestive (b) (i) protists (ii) transcription (c) (i) plant cells (ii) replication (d) (i) prokaryotes (ii) defence (ii) Which of the following statements regarding modification enzyme is correct?
(a) It adds methyl group to one or two bases usually within the host DNA sequence to protect it from the restriction enzyme. (b) It adds ethyl group to one or two bases usually within the sequence recognised by the restriction enzymes. (c) It adds methyl group to only one of bases within the foreign DNA sequence that is recognised by the restriction enzymes. (d) None of these (iii) Which of the following is a type II restriction enzyme?
(a) Alu I (b) EcoR I (c) BamH I (d) All of these (iv) Which of the following is the first discovered restriction endonuclease?
(a) Sal I (b) EcoR I (c) Hind II (d) EcoR II (v) Components of Restriction Modification System include
(a) restriction enzyme (b) modification enzyme (c) lysing enzyme (d) both (a) and (b). (a) -
Read the following and answer any four questions from (i) to(v) given below:
In recombinant DNA technology, the fragments of DNA generated after cutting the DNA by restriction enzymes are separated according to their size or length by gel electrophoresis. Gel electrophoresis is performed in a gel matrix so that molecules of similar electric charges can be separated on the basis of size. Most commonly used matrix in gel electrophoresis is agarose. The fragments are separated under the influence of electric field. The separated DNA fragments can be seen only after staining the DNA with compound known as ethidium bromide (EtBr) followed by exposure to UV radiation as bright orange band.
(i) Gel electrophoresis is used for the separation of(a) DNA only (b) DNA and RNA only (c) DNA and proteins only (d) DNA, RNA and proteins. (ii) Most commonly used matrix is (i) which is a (ii) extracted from (iii).
(a) (i) agarose (ii) polysaccharide (iii) sea weed (b) (i) agarose (ii) protein (iii) sea weed (c) (i) EtBr (ii) polysaccharide (iii) sea weed (d) (i) EtBr (ii) protein (iii) bacteria (iii) A DNA molecule was treated with a restriction endonuclease and three fragments of size (i) 426 kb, (ii) 129 kb and (iii) 46 kb were obtained. Identify the order in which these bands will arrange themselves in the gel plate after gel electrophoresis is completed. (Assuming that negative part of electrode is towards the well)
(a) (iii) ➝ (ii) ➝ (i) (b) (i) ➝ (ii) ➝(iii) (c) (i) ➝ (iii) ➝ (ii) (d) (iii) ➝ (i) ➝ (ii) (iv) Which of the following statements regarding gel electrophoresis is incorrect?
(a) Separated DNA fragments can be seen only after staining DNA with EtBr. (b) DNA fragments are separated according to their size. (c) Under the influence of electric field, positively charged molecules move towards the anode and negatively charged molecules move towards the cathode. (d) None of these (v) The factor that will not affect the rate of DNA migration in gel electrophoresis is
(a) size of DNA molecule (b) concentration of DNA (c) voltage supplied (d) concentration of the gel (a)
Case Study
*****************************************
Answers
Biotechnology: Principles and Processes Case Study Questions With Answer Key Answer Keys
-
(a) Recombinant DN Atechnology/Genetic engineering.
(b) 1. Isolation of the desired segment of DNA.
2. Ligation of the isolated DNA to a plasmid vector.
3. Introduction of rDNA into the bacterial cell.
4. The bacterium produces the gene product.
(c) Human insulin is produced by transgenic Escherichia coli cells. -
(a) Restriction endonuclease - the same restriction enzyme cuts the foreign DNA and vector/plasmid DNA.
(b) DNA ligase joins the foreign DNA segment to the plasmid.
(c) A is transformation.
B is cell division or clone formation. -
(a) It is called a palindromic sequence.
(b) The overhanging chains, called 'sticky ends' facilitate the action of DNA ligase.
(c) EcoRI. -
(i) a–Vector DNA
b–Foreign DNA
(ii) 5' — GAATTC — 3'
3' — CTTAAG — 5'
(iii) DNA ligase -
(a) It depicts agarose gel electrophoresis used for separation of DNA fragments.
(b) The 'largest' and 'smallest' mean the DNA fragments oflarge size and small size, respectively.
(c) Ethidium bromide.
(d) Elution is the process in which the separated bands of DNA are cut out from the gel and extracted. -
(a) A is the negative terminal.
B is the positive terminal.
(b) Agarose is the matrix; it is obtained from sea weeds.
(c) C is the undigested DNA. -
(a) G - Ampicillin-resistance gene
H- Tetracycline-resistance gene
(b) A - BamHl, B - Sal I
(c) F is EcoRI
(d) Ori. -
(a) A - Denaturation; D - Extension.
(b) B - Primers; they are oligonucleotides.
(c) C - Taq polymerase; its source organism is Thermus aquaticus. -
(a) It is a simple stirred-tank bioreactor.
(b) The curved base facilitates the mixing of the contents in the reactor.
(c) The stirrer facilitates even mixing and oxygen availability throughout the bioreactor. -
(a) In this method, the cells are maintained in their physiologically most active exponential phase; there is production of a larger biomass and hence, higher yield of the gene product.
(b) The optimal conditions include
(i) temperature,
(ii) pH,
(iii) oxygen,
(iv) substrate concentration. -
(i) (d)
(ii) (b) : Plasmid in a bacterial cell is an extra chromosomal material that undergo replication with or without chromosomal DNA.
(iii) (a)
(iv) (c) : In 1972, Paul Berg was able to introduce a gene of SV-40 virus into a bacterium with the help of lambda phage.
(v) (b) -
(i) (d) : Only type II restriction enzymes are used in gene manipulation for two reasons:
(a) No ATP is required for the cleaving action.
(b) It makes cleavage or cut in both the strands of DNA molecule.
(ii) (a) : All the three types of restriction endonucleases require Mg2+ ions for restriction.
(iii) (c) : Restriction endonuclease was isolated for the first time by W Arber in 1962 in bacteria.
(iv) (b)
(v) (b) -
(i) (a) : Conversion of starch into maltose and protein into amino acids is due to hydrolysis.
(ii) (c) : DNA polymerase I
(iii) (b)
(iv) (c)
(v) (c): Type I restriction endonuclease consist of three different subunits and requires ATP, Mg2+, S-adenosyl methionine for restriction. -
(i) (d)
(ii) (a)
(iii) (d) : Different examples of Type II restriction endonuclease are Alu I, EcoR I, BamH I, etc.
(iv) (c)
(v) (d) : The restriction modification system consists of two components
(i) A restriction enzyme called restriction endonucleases which identifies the introduced foreign DNA and cuts it into pieces and
(ii) A modification enzyme (methylase) that adds a methyl group to DNA at a specific site to protect the site from restriction endonuclease cleavage. -
(i) (d) : Gel electrophoresis is a technique used to separate fragments of molecules, i.e., DNA, RNA and protein.
(ii) (a)
(iii) (b)
(iv) (c) : Under the influence of electric field positively charged molecules move towards the cathode and negatively charged molecules move towards the anode.
(v) (b) : Concentration of DNA will not affect the migration of DNA molecule in a gel electrophoresis. As in gel electrophoresis, molecules separate according to their size therefore DNA size will affect migration. Increased voltage supply will increase rate of migration and the more concentrated gel will reduce rate of migration.